by: Agnes Purwidyantri
Wine fraud, including counterfeiting labels and brands, adulteration using unauthorised additives or practices, and substitution based on grape variety or region of origin counterfeits and mislabelling, has threatened wine producers and policymakers globally. Authentic and high-quality wine is controlled and produced under a set of legislative frameworks, namely Geographical Indications (GI) recognition, namely Protected Designation of Origin (PDO), Protected Geographical Indication (PGI) (Regulation (EU) No 1308/2013), and Geographical Indications (GI) of spirits drinks and aromatised wines. The quality monitoring and authenticity assessment of wines typically rely on analytical and chemo metric methods. The target of analysis varies from mainly chemical components in wine, for instance, the level of molecular organic acids, volatile compounds, polyphenols, amino acids, biogenic amines, and inorganic species that are generally climatic, geographical, and technical1. To date, wine analyses employ classical gas chromatography-mass spectrometry (GC-MS) and high-performance liquid chromatography (HPLC) techniques. Some other detections using atomic absorption spectrometry (AAS) and inductively coupled plasma—mass spectrometry (ICP-MS) or combinations of these techniques have also been applied in mineral tracing. More recently, FTIR (Fourier Transform Infrared Spectroscopy) spectrometry and NMR (Nuclear Magnetic Resonance) spectroscopy have been widely used in wine authenticity studies2 . These methods have shown great applicability and robust results suitable for industrial settings. However, they come with complex statistical analyses of an extensive data set, bulky and pricey instruments, inapplicable and inaccessible for grape growers and small-scale wineries.

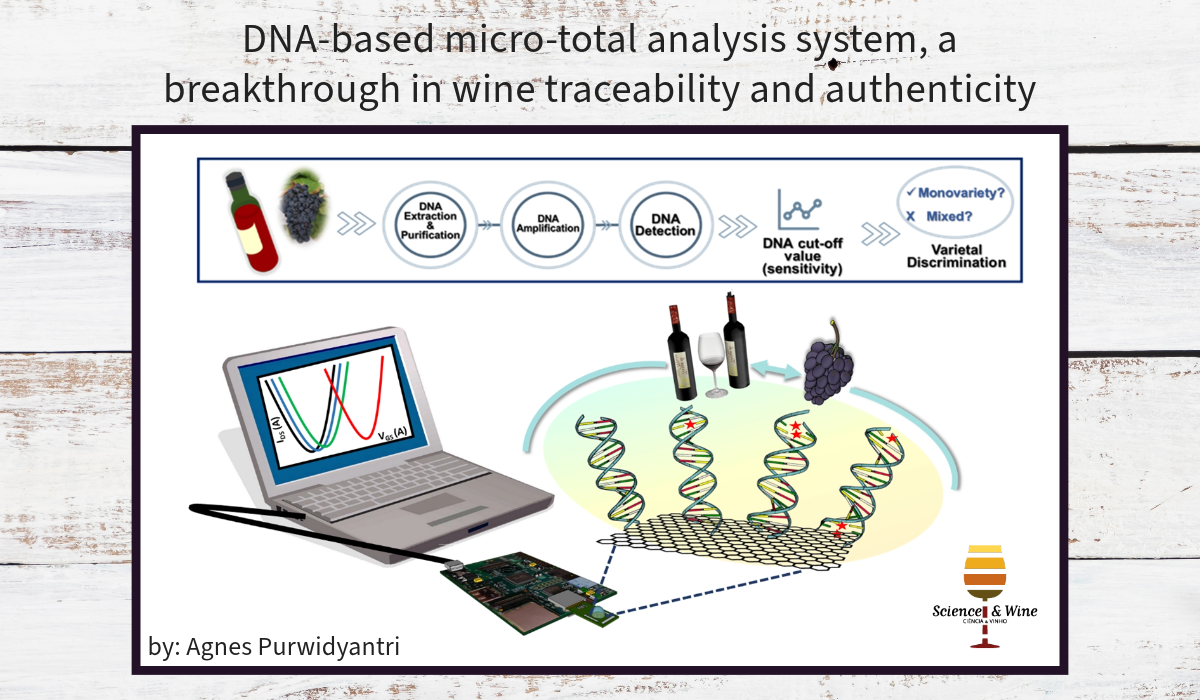
DNA-based analysis to trace specific grapevine varieties used in the winemaking process provides several advantages over the existing methods because of the following factors. First, unlike the metabolic components of wine mentioned above, DNA is highly stable, even under harsh environments, seasonal or climate factors, and a long period of evolution. Second, single nucleotide polymorphism (SNP) refers to changes in a single base at a specific position in the genome that could be a robust biomarker to distinguish grapevine varieties. Some well-known genes with unique SNPs locations have been objects of investigation in wine control, such as F3H related to anthocyanin pathways and LDOX, the direct precursor enzyme of most of the flavonoid classes 3. The main challenges in DNA-based analysis arise from the low content and purity of DNA in wine. Therefore, an integrated strategy that provides solutions for effective DNA extraction, amplification, purification, and accurate varietal discrimination is urgently required.
Micro total analysis systems (µTAS) and lab-on-a-chip (LoC)-based biosensors have been game changers in DNA analysis and are of great interest in the wine industry. Incorporating microfluidic technology and advanced materials paves an exciting avenue toward rapid, sensitive, highly portable, and cost-effective methods for vast applications, from sample preparation to detection. In the DNA extraction stage, microfluidic chips fabricated using low-cost materials, such as Polydimethylsiloxane (PDMS) and Poly (methyl methacrylate) (PMMA), through easy 3D printing methods, have drawn significant attention from industry and research 4,5. Combining and optimising the micro-solid phase extraction (µSPE) techniques in a microfluidic system could be a versatile route toward effective DNA yield and purity 6. The easy, handy, and user-friendly microfluidic extraction kit is the potential for mass production. It is also suitable for on-site wine DNA extraction, which could be an alternative to expensive and laborious commercial DNA extraction kits. Subsequently, integrating a DNA amplification module into a microfluidic setup enables single DNA strand amplification and DNA concentration. Besides polymerase chain reaction (PCR)-based, (Loop-Mediated Isothermal Amplification) LAMP could be manifested on-chip and combined with simple detection methods, such as colorimetry, that even allow rapid and naked-eye detection 7.

The ultimate module in an integrated system is the DNA detection part. Graphene field-effect transistors (FETs) exhibit excellent parameters for ultrasensitive DNA detection among myriads of advanced materials and sensing devices. FETs present direct and label-free detection and are easily integrated with commercially available data analysers and transducers. The underlying principle of FET is in the conductivity’s change of the interface as the biochemical reaction, for instance, DNA hybridisation, occurs. Integrated into an FET system, graphene, with its one-atom molecule thickness and remarkably high conductivity and mobility, has been the best candidate as a bio-recognition element in DNA detection. Importantly, graphene is also biocompatible and easily functionalised. Notably, among other 2D materials fabrication, only graphene classes, including graphene films and graphite oxide powders, are commercially available and verified for wafer-scale production. For miniaturisation and prototyping, the graphene FET system could be assembled with programmable microfluidics allowing automatization in DNA hybridisation study 8. In a study conducting varietal discrimination from grapevine and wine samples using SNPs, the graphene FET sensor demonstrated an ultrasensitive detection down to the attomolar with the ability to distinguish low-level mismatches and the possibility for multiplex screening. The outstanding performance of graphene FET for µTAS offers a ground-breaking technology for a rapid, automated, portable, and deployable system for decentralised wine authenticity control and assessment.
Read more at:
Integrated Approach from Sample-to-Answer for Grapevine Varietal Identification on a Portable Graphene Sensor Chip. Agnes Purwidyantri*, Sarah Azinheiro, Aitor García Roldán, Tereza Jaegerova, Adriana Vilaça, Rofer Machado, M. Fátima Cerqueira, Jérôme Borme, Telma Domingues, Marco Martins, Pedro Alpuim*, and Marta Prado. ACS Sensors. 2023. https://pubs.acs.org/doi/10.1021/acssensors.2c02090

Agnes Purwidyantri
Agnes is currently a research fellow at Queen’s University Belfast, UK. She has extensive experience of over 10 years in multidiscipline research and multinational environment. Previously, she worked as a research fellow at INL Portugal and was involved in developing a micro-total analysis system (µTAS) and graphene sensors for a wide range of applications, such as food and beverage fraud control, biomedical and environmental screening. She previously served as scientist the National Research and Innovation Agency of Indonesia/ BRIN) at Research Unit for Clean Technology. She was a postdoctoral fellow at Biosensor Group, Chang Gung University, Taiwan, and worked on Au nanostructure for multi-implemented sensors and high-k materials for biomedical projects involving Chang Gung Memorial Hospital and Taiwan Semiconductor and Manufacturing Company (TSMC). Agnes received her Ph.D. degree in Biomedical Engineering from Chang Gung University, Taiwan in 2017, with thesis work on the development of DNA sensors for kidney disease. Her master’s degree was in Biotechnology and was received in 2013 from Asia University, Taiwan, where she worked to exploit the enrichment of medicinal fungi through pulsed-UV light exposure. She obtained her bachelor’s in food technology in 2007 from Soegijapranata Catholic University, Indonesia, with a project investigating corn rice’s physicochemical and organoleptic properties. She is Review Editor of Frontiers in Food Science and Technology, Associate Editor of IEEE Open Journal of Nanotechnology (OJ-NANO), Guest Editor for MDPI Electronics-Special Issue on Nanosensors: Sensing Principle, System and Application of Electronics, and Special Issue on Embedded FET for Application as a Biosensor.
References.
(1) Sun, X.; Zhang, F.; Gutiérrez-Gamboa, G.; Ge, Q.; Xu, P.; Zhang, Q.; Fang, Y.; Ma, T. Real Wine or Not? Protecting Wine with Traceability and Authenticity for Consumers: Chemical and Technical Basis, Technique Applications, Challenge, and Perspectives. Critical Reviews in Food Science and Nutrition. 2022. https://doi.org/10.1080/10408398.2021.1906624.
(2) Pereira, L.; Gomes, S.; Barrias, S.; Gomes, E. P.; Baleiras-Couto, M.; Fernandes, J. R.; Martins-Lopes, P. From the Field to the Bottle—an Integrated Strategy for Wine Authenticity. Beverages. 2018. https://doi.org/10.3390/beverages4040071.
(3) Gomes, S.; Castro, C.; Barrias, S.; Pereira, L.; Jorge, P.; Fernandes, J. R.; Martins-Lopes, P. Alternative SNP Detection Platforms, HRM and Biosensors, for Varietal Identification in Vitis Vinifera L. Using F3H and LDOX Genes. Sci Rep 2018, 8 (1), 1–12. https://doi.org/10.1038/s41598-018-24158-9.
(4) Anshori, I.; Lukito, V.; Adhawiyah, R.; Putri, D.; Harimurti, S.; Rajab, T. L. E.; Pradana, A.; Akbar, M.; Syamsunarno, M. R. A. A.; Handayani, M.; Purwidyantri, A.; Prabowo, B. A. Versatile and Low-Cost Fabrication of Modular Lock-and-Key Microfluidics for Integrated Connector Mixer Using a Stereolithography 3D Printing. Micromachines (Basel) 2022, 13 (8). https://doi.org/10.3390/mi13081197.
(5) Hassanpour-Tamrin, S.; Sanati-Nezhad, A.; Sen, A. A Simple and Low-Cost Approach for Irreversible Bonding of Polymethylmethacrylate and Polydimethylsiloxane at Room Temperature for High-Pressure Hybrid Microfluidics. Sci Rep 2021, 11 (1). https://doi.org/10.1038/s41598-021-83011-8.
(6) Carvalho, J.; Puertas, G.; Gaspar, J.; Azinheiro, S.; Diéguez, L.; Garrido-Maestu, A.; Vázquez, M.; Barros-Velázquez, J.; Cardoso, S.; Prado, M. Highly Efficient DNA Extraction and Purification from Olive Oil on a Washable and Reusable Miniaturized Device. Anal Chim Acta 2018, 1020, 30–40. https://doi.org/10.1016/j.aca.2018.02.079.
(7) Azinheiro, S.; Roumani, F.; Prado, M.; Garrido-Maestu, A. Rapid Same-Day Detection of Listeria Monocytogenes, Salmonella Spp., and Escherichia Coli O157 by Colorimetric LAMP in Dairy Products. Food Anal Methods 2022, 15 (11), 2959–2971. https://doi.org/10.1007/s12161-022-02345-9.
(8) Purwidyantri, A.; Ipatov, A.; Domingues, T.; Borme, J.; Martins, M.; Alpuim, P.; Prado, M. Programmable Graphene-Based Microfluidic Sensor for DNA Detection. Sens Actuators B Chem 2022, 367. https://doi.org/10.1016/j.snb.2022.132044.